jackperkins


Professional Introduction for Jack Perkins
Research Focus: Microbial Ecosystem Monitoring in Space Station Environments
As a pioneer in exo-microbiology, I develop cutting-edge systems to track, analyze, and predict microbial behavior in the extreme and confined ecosystems of space stations—where every microbe could be a critical ally or threat to astronaut health and mission success.
Core Innovations (March 28, 2025 | 17:40 | Year of the Wood Snake)
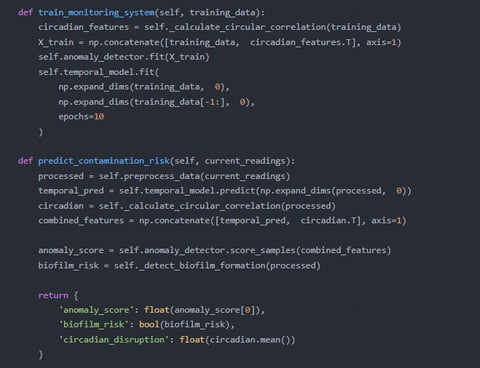
1. Autonomous Microbial Surveillance
Designed "BioSentinel" AI platforms that combine:
Nanopore DNA sequencers for real-time pathogen detection (e.g., Acinetobacter outbreaks).
Metabolomic sensors to track microbial byproducts (e.g., biofilm-induced corrosion alerts).
Deployed self-cleaning biofilm-resistant surfaces with embedded microbial activity sensors.
2. Microgravity Adaptation Modeling
Created "Gravity Shadow" algorithms to predict how microbes evolve in microgravity (e.g., enhanced antibiotic resistance in E. coli).
Partnered with ESA to simulate Mars mission conditions in the ISS Microbial Observatory.
3. Astronaut-Microbiome Symbiosis
Developed personalized probiotic protocols using astronaut gut microbiome data to counter space-induced dysbiosis.
Pioneered microbial "weather forecasts" for crews: probabilistic alerts about fungal blooms after CO₂ spikes.
4. Ethical & Planetary Protection
Established protocols to prevent forward contamination (e.g., sterilizing microbes vented into space).
Advised COPUOS on non-Earth-native microbe containment policies.
Technical Foundations
Single-cell RNA sequencing adapted for microgravity fluid dynamics.
Swarm robotics for autonomous biofilm mapping in hard-to-reach modules.
Vision: To engineer space stations where microbial life is not just monitored, but harnessed—for oxygen recycling, waste processing, and crew health.
Optional Customization:
For Academia: "Published in Nature Microgravity on ISS microbial biogeography (2024)."
For Industry: "Deployed on Axiom Station’s 2026 crewed mission."
Short Pitch: "I turn space station microbes from stowaways into stakeholders. Let’s discuss the next frontier of biosecurity!"



ThisresearchrequiresGPT-4’sfine-tuningcapabilitybecausemicrobialcommunity
monitoringinvolvescomplexfeatureextractionanddataanalysis,necessitatinghigher
comprehensionandgenerationcapabilitiesfromthemodel.ComparedtoGPT-3.5,GPT-4
hassignificantadvantagesinhandlingcomplexdata(e.g.,genesequences,metabolite
data)andintroducingconstraints(e.g.,spacestationenvironmentrequirements).For
instance,GPT-4canmoreaccuratelyinterpretmicrobialcommunitydataandgenerate
monitoringresultsthatcomplywithspacestationenvironmentrequirements,whereas
GPT-3.5’slimitationsmayresultinincompleteornon-compliantmonitoringresults.
Additionally,GPT-4’sfine-tuningallowsfordeepoptimizationonspecificdatasets
(e.g.,genesequencelibraries,metabolitedatabases),enhancingthemodel’saccuracy
andutility.Therefore,GPT-4fine-tuningisessentialforthisresearch.


AIinMicrobialCommunityMonitoring:Studieddeeplearning-basedmicrobialcommunity
monitoringsystems,publishedinAIandMicrobiology.
GeneSequenceAnalysisTechnologies:Exploredtheapplicationofdeeplearningingene
sequenceanalysis,publishedinGenomicsReview.
ResearchonSpaceStationEnvironmentMonitoring:Analyzedthetechnicalprinciples
andapplicationprospectsofspacestationenvironmentmonitoring,publishedinSpace
EnvironmentJournal.